 putEMG and putEMG-Force datasets are databases of surface electromyographic activity recorded from forearm. Datasets allows for development of algorithms for gesture recognition and grasp force recognition. Experiment was conducted on 44 participants, with two repetitions separated by, minimum of one week. The dataset includes 7 active gestures (like hand flexion, extension, etc.) + idle and a set of trials with isometric contractions. sEMG was recorded using a 24-electrode matrix.
putEMG and putEMG-Force datasets are databases of surface electromyographic activity recorded from forearm. Datasets allows for development of algorithms for gesture recognition and grasp force recognition. Experiment was conducted on 44 participants, with two repetitions separated by, minimum of one week. The dataset includes 7 active gestures (like hand flexion, extension, etc.) + idle and a set of trials with isometric contractions. sEMG was recorded using a 24-electrode matrix.
How can I use the putEMG and putEMG-Force dataset?
- Follow the description in Download section, download manually or use automated scripts.
- Choose your preferred file flavour (HDF5 or CSV) and familiarize yourself with data formats and code examples in Data format section.
- Evaluate your algorithms using putEMG and putEMG-Force datasets. See Examples with putEMG section for easy start.
- When using putEMG datasets please cite the corresponding paper listed below:
When using putEMG: sEMG gesture recognition dataset:
Kaczmarek, P.; Mańkowski, T.; Tomczyński, J. putEMG—A Surface Electromyography Hand Gesture Recognition Dataset. Sensors 2019, 19, 3548.
@article{putEMGKaczmarek2019,
author = {Kaczmarek, Piotr and Mańkowski, Tomasz and Tomczyński, Jakub},
title = {putEMG—A Surface Electromyography Hand Gesture Recognition Dataset},
journal = {Sensors},
volume = {19},
year = {2019},
number = {16},
article-number = {3548},
url = {https://www.mdpi.com/1424-8220/19/16/3548},
issn = {1424-8220},
doi = {10.3390/s19163548}
}
When using putEMG-Force: sEMG grasp force estimation dataset: TBD
Download
If you would like to download putEMG datasets please use the link below. Each folder contains a MD5SUMS files with MD5 checksums for each file in folder:
You can either download the data manually with browser using the link above, or use an automated bash or Python (3.6) download scripts provided in git repository:
https://github.com/biolab-put/putemg-downloader
To download all experiment data in HDF5 format along with 1080p video, depth images, please issue following commands (for other data formats, see README file or run ./putemg_downloader.py for usage):
git clone https://github.com/biolab-put/putemg-downloader.git cd putemg-downloader # Python 3.6 ./putemg_downloader.py emg_gestures,emg_force data-hdf5,video-1080p,depth # Bash + Wget ./putemg_downloader.sh emg_gestures,emg_force data-hdf5,video-1080p,depth
Participants
The dataset includes 44 healthy subjects (8 females, 36 males) aged 19 to 37 years old. Each subject participated in the experiment twice, with a minimum one week interval, performing same procedure.
Each subject signed a participation consent with data publication permit. The study was approved by Bioethical Comitee of Poznan University of Medical Science under no 398/17. Anonymous participant list, along with health condition questionnaire can be found here:
PARTICIPANTS SUMMARY: CSV , XLSX
Experiment Platform
A workbench dedicated to sEMG signal acquisition was developed. The system allows for forearm muscle activity recording for a single subject. The acquisition platform is visible below:
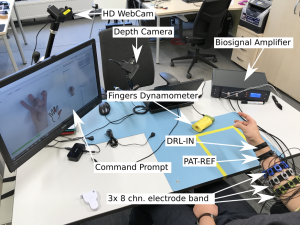
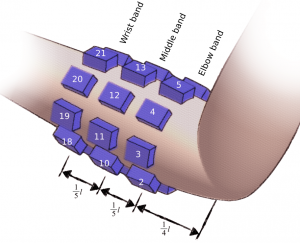
Signals are recorded from 24 electrodes fixed around subject right forearm using 3 elastic bands, creating a 3×8 matrix. Electrodes were evenly spaced (45°) around participant’s forearm. First band was placed in aprox. 1/4 of forearm length measuring from the elbow, each following band was separated by aprox. 1/5 of forearm length. First electrode of each band was placed over the ulna bone and numbered clockwise respectively. Resulting in following numbering pattern: elbow band [1-8], middle band [9-16], wrist band [17-24]. Each electrode is a 10 mm Ag/AgCl-coated element in a 3D-printed housing. Proper skin contact was assured with sponge insert saturated with electrolyte. Different band sets were used in order to compensate participants forearm diameter differences.
 |
 |
Main acquisition hardware is a desktop multichannel EMG amplifier – MEBA by OT Bioelettronica. The data was sampled at 5120 Hz, with 12-bit A/D conversion using 3 Hz high-pass and 900 Hz low-pass filter. All signals were pre-amplified with gain of 5, using amplifiers placed on subject’s arm, resulting in total gain of 200. Signals were recorded in monopolar mode with DRL-IN and Patient-REF electrodes placed in close proximity of the wrist of examined arm.
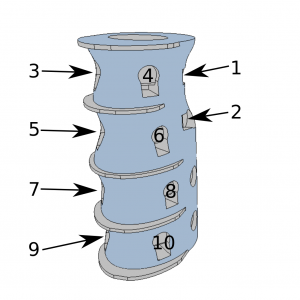
In order to provide a possibility of isometric gesture/contraction assessment, putEMG Dataset includes trials where not gesture but force exerted by particular fingers was required as an input. For that purpose, an ergonomic hand dynamometer was developed. For each finger, the dynamometer features 2 tensometer sensor points and analogue linearisation circuit. The sensor was shaped ergonomically allowing fingers separation. Data is sampled at 200 Hz rate. Before each experiment, sensor bias is subtracted. Force data is interpolated to match EMG sampling rate
 |
 |
For gesture execution ground-truth, a HD Camera providing an RGB feed and a depth camera with a close view of subject’s hand were used. Videos and depth images are provided alongside the EMG data.
Proprietary PC software was used to prompt the participant a task to be performed. The software operated in two modes: gesture and force. In gesture mode, the subject was presented with a photo and pictogram of desired gesture. Next gesture preview and countdown to gesture transition was also displayed. In force mode, desired trajectory is shown to the participant in a scrolling timeline, and the subject tries to match current force data of each finger with given trajectory.
Experiment Description
Experiment was performed twice for each subject, with at least one week separation. Experiment consisted of two main parts: gesture (where subject was asked to repeat given hand gesture) and force (where force trajectory of each finger’s isometric flexion was given). Participants were asked to sit comfortable in a chair and rest the elbow on an armrest. For both gesture and force experiments, participants performed a familiarization trajectory, which is not included in the Dataset.
In case of gesture trajectories participants were asked to hold the forearm with inclination of 10-20°. Gesture set includes the following 8 gestures: idle, fist, flexion, extension and 4 pinches (full description and pictures below). Each active gesture was separated by a 3-second idle period. Each time, active gesture, was held for 1 or 3 seconds. Trajectories were divided into action blocks between which the subject could relax and move hand freely. Gesture experiment consisted of three trajectories (resulting in 20 repetitions of each active gesture):
- repeats_long – 7 action blocks, each block contains 8 repetitions of each active gesture;
- sequential – 6 action blocks, each block is an subsequent exectution of all active gestures;
- repeats_short – 7 action blocks, each block contains 6 repetitions of each active gesture.
During force trajectories, the participants were asked to firmly grasp the dynamometer, and rest the device on the table. Before each force experiment, tensometer bias was acquired and subtracted. Then, maximal voluntary contraction (MVC) force level was recorded (included in the Dataset). Magnitudes of force trajectories were then scaled to fit particular subject’s MVC. All trajectories contain finger presses separated by 3-second idle state. Longer pauses between large action blocks were also provided for muscle relaxation. Trajectories consist of varying combinations of pressure magnitude, duration and shape (step and ramp). Trajectories include presses of: thumb, index, middle, ring+small and all fingers. Force experiment consisted of five trajectories:
- bias – force sensor bias acquisition trajectory, from this trajectory a sensor bias is extracted, and subtracted from each succeeding force trajectories;
- mvc – contains only one step, when subject was asked to squezze the dynamometer as hard as possible;
- repeats_long – 5 action blocks, each press type repeated 11 times with varying parameters;
- sequential – 4 action blocks, each block is a repeated two times subsequent execution of each press type, with varying parameters;
- repeats_short – 5 action blocks, each press type repeated 5 times with varying parameters.
Preprocessing
Raw EMG signal was not altered by any means of signal processing. Each trial is an uninterrupted signal stream that includes both static parts and transitions. All trials are labelled by a given gesture or force trajectory, however gesture/press execution may not perfectly match the trajectory label. For force experiment, ground-truth is provided by dynamometer measurement, interpolated (forward hold) to match EMG sampling. In case of gesture experiment additional ground-truth label is provided by hand pose estimation using video stream and deep neural network.
Data Formats
putEMG Dataset comes in two file formats: HDF5 and CSV (zipped for convenient size). When using HDF5 format we recommend using pandas – Python Data Analysis Library:
Python
import pandas as pd
data = pd.read_hdf('file_path/trial_name.hdf5')
For Matlab/GNU Octave environment CSV file (after unzipping) is recommended:
Matlab
data = csvread('file_path/trial_name.csv', 1, 0);
header = strsplit(fgetl(fopen('file_path/trial_name.csv')), ',')
Each trial name has the following format:
<type>-<subject>-<trajectory>-YYYY-MM-DD-hh-mm-ss-milisec, where:
- <type> – defines experiment type: emg_gesture | emg_force
- <subject> – two digit participant identifier
- <trajectory> – defines trajectory type: mvc | repeats_long | sequential | repeats_short
emg_gesture experiment files include following columns:
- Timestamp
- EMG_1 … EMG_24 – measured sEMG ADC value, labeled as in Experiment Platform;
- TRAJ_1 – label of gesture shown to the subject;
- TRAJ_GT_NO_FILTER – video stream gesture estimation, without processing;
- TRAJ_GT – label of gesture estimated from video stream using DNN (ground-truth), after processing – use this column as final groud-truth for gesture classification;
- VIDEO_STAMP – sample timestamp in corresponding video stream;
emg_force experiment files include following columns:
- Timestamp
- EMG_1 … EMG_24 – measured sEMG ADC value, labeled as in Experiment Platform
- FORCE_1 … FORCE_10 – measured force value (ground-truth) of each tensometer, labeled as in picture;
- FORCE_MVC – MVC value measured during mvc trajectory;
- TRAJ_1 … TRAJ_4 – force trajectories that the subject had to follow, with following order: thumb, index, middle, ring+small;
EMG_1 … EMG_24 is stored as raw ADC data, in order to calculate voltage use the following formula, where N is an ADC value:
![]()
Additionally all HDF5 files include pandas.Categorical columns marking all samples with: type, subject, trajectory and date_time extracted from trial name.
All trials are accompanied by webcam video stream in both 1080p and 576p resolution in H264 MP4 format. Video stream was used to estimate gesture execution and can be used to verify experiment course. Video file names are same as corresponding trial names.
Data from depth camera is available for emg-gesture experiments in PNG format with resolution of 640×480 px. PNG files are named depth_<timestamp>.png, with timestamp in time domain of corresponding trial file, in miliseconds. Depth files are stored in a zip archive with name the same as corresponding trial.
Examples with putEMG
Dedicated exemplary Python3 scripts to be used with putEMG dataset are available at following git repository:
https://github.com/biolab-put/putemg_examples
Contact
If you have any questions concerning putEMG datasets feel free to contact us using info on Contact page. We will be happy to help to solve any problem with download, file formats, data evaluation, benchmarking etc. and provided any additional information.
License
Unless stated otherwise, all putEMG datasets elements are licensed under a Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0). If you wish to use putEMG datasets in commercial project please contact our team via Contact page. Accompanying scripts are licensed under a MIT License.
Acknowledgements
This work was supported by a grant from Polish National Science Centre, project PRELUDIUM 9, research project no. 2015/17/N/ST6/03571.









